 Maintaining a high-fidelity proteome is essential for the health of cells and organisms. Producing thousands of proteins with diverse functions requires highly coordinated multi-step biosynthetic pathways that are tightly integrated with quality control mechanisms. To function properly, nascent proteins must be accurately synthesized from mRNAs, fold into specific three-dimensional structures, undergo posttranslational modifications, assemble into multi-component complexes, and correctly localize to the appropriate cellular destination. Each step of protein biosynthesis is susceptible to errors that must be recognized by quality control mechanisms and either corrected or degraded. In addition, many protein biosynthetic pathways are closely monitored and regulated to support physiological processes and prevent disease. Our lab identifies and dissects molecular mechanisms that determine the fates of nascent proteins using strategies that incorporate biochemistry, cell biology, and structural biology.
Maintaining a high-fidelity proteome is essential for the health of cells and organisms. Producing thousands of proteins with diverse functions requires highly coordinated multi-step biosynthetic pathways that are tightly integrated with quality control mechanisms. To function properly, nascent proteins must be accurately synthesized from mRNAs, fold into specific three-dimensional structures, undergo posttranslational modifications, assemble into multi-component complexes, and correctly localize to the appropriate cellular destination. Each step of protein biosynthesis is susceptible to errors that must be recognized by quality control mechanisms and either corrected or degraded. In addition, many protein biosynthetic pathways are closely monitored and regulated to support physiological processes and prevent disease. Our lab identifies and dissects molecular mechanisms that determine the fates of nascent proteins using strategies that incorporate biochemistry, cell biology, and structural biology.
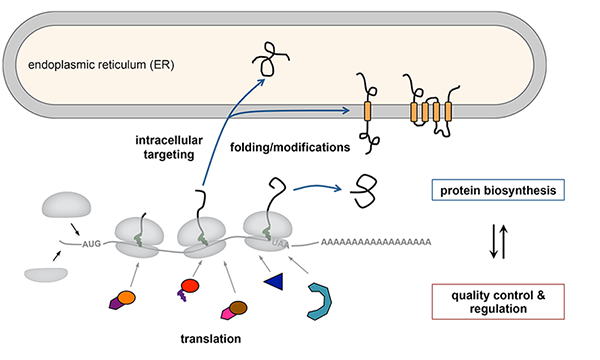
Topics we study:
- Quality control of nascent protein synthesis
- Molecular recognition of aberrant translation
- Ribosome-associated protein quality control
- Regulation of translation termination
- Function and regulation of ribosome-associated factors
- Membrane protein biogenesis
- Protein sorting to different cellular organelles
- Membrane protein quality control
- Regulation of membrane protein abundance
- Membrane protein topogenesis
Our approaches include:
- Reconstituting protein biosynthesis and quality control processes using cell culture, in vitro lysate-based, and completely purified experimental systems.
- Developing methods and generating reagents to identify, uncouple, and mechanistically dissect the individual steps and molecular components of these cellular pathways.
- Isolating or assembling biologically-relevant macromolecular complexes that represent functional intermediates of these pathways for structural analysis, primarily via cryogenic electron microscopy (cryo-EM).

